Invited by Prof. Chen Song, Prof. Wonpil lm from Lehigh University visited us and presented his groundbreaking work on the biomolecular simulation program CHARMM-GUI. CHARMM-GUI is a widely-used software suite that facilitates the setup, simulation, and analysis of a diverse range of biomolecular systems, including proteins, lipids, and nucleic acids. In his presentation, Prof. Wonpil lm highlighted the exceptional capabilities and versatile applications of this powerful computational biology tool, which has become an invaluable resource for the global research community.
Prof. Shengyou Huang's visit
SLC19A3 paper online
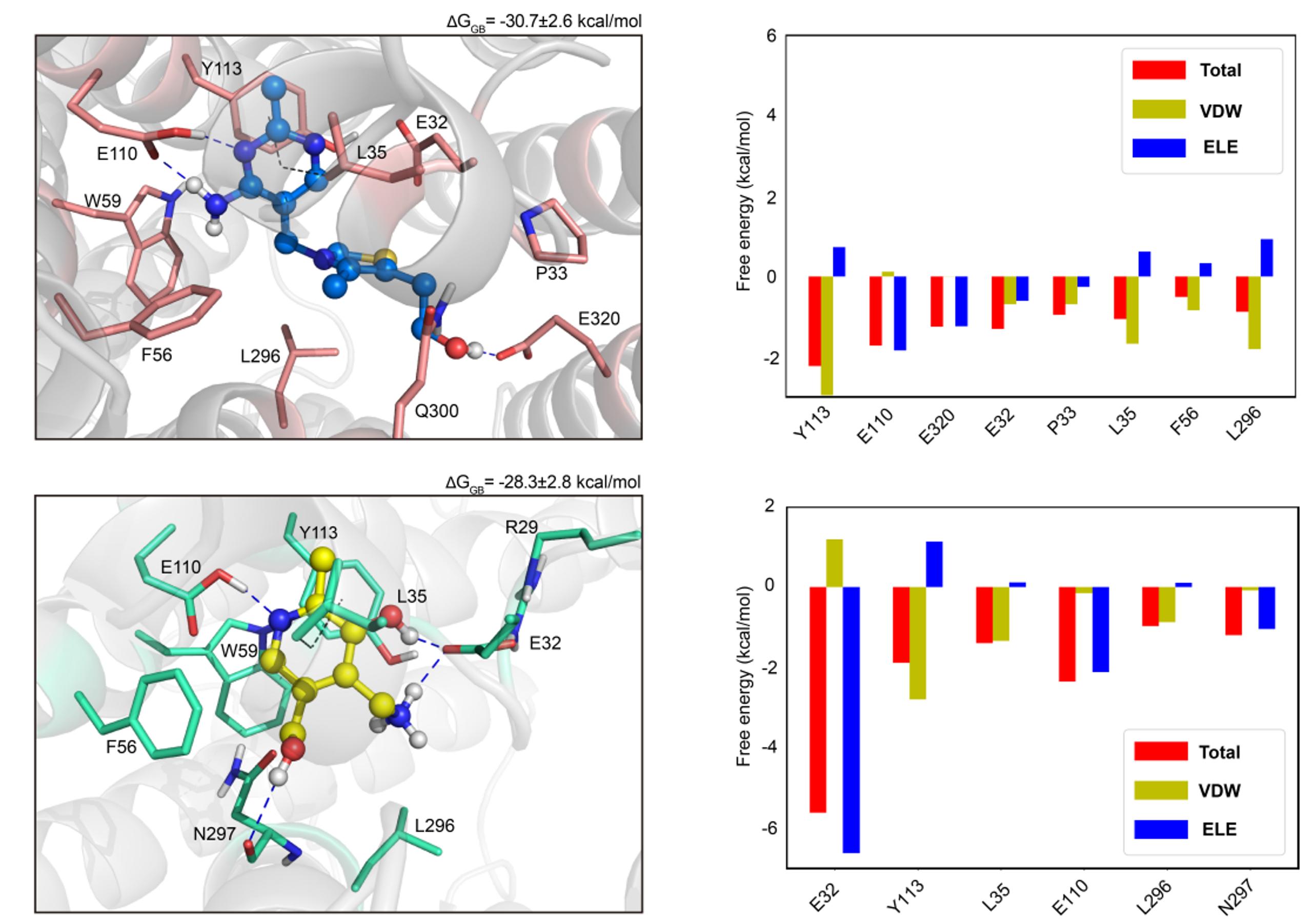
The collaborative project focused on SLC19A3 has been published in Cell Research online! The study was conducted in partnership with Prof. Zhe Zhang's lab, and we employed molecular dynamics (MD) simulations and binding free energy calculations to help determine the complex structures of human thiamine transporter 2 (ThTr2 or SLC19A3). Congratulations to Nanhao on the successful publication of this research!
Switching Gō-Martini Paper Online
YANG Song's first paper detailing the development of the Switching Gō-Martini method has been published online in JCTC (Link). This method promises to be a valuable tool for efficiently simulating the likely conformational transitions between two structures of a protein. When used in conjunction with Jiaxuan's Alternative Conformation Modeling approach (Link), in principle, one can begin with a protein sequence, predict two probable conformations, and then simulate the potential transition pathways between them.
CQB Annual Meeting 2023
2023 has been an extraordinary year, marked by significant achievements and numerous accolades for our group members. Qingyang, Jiaxuan, Zefeng, Ruihan, Kai, and Lingfeng have each secured various scholarships, and Song distinguished himself by earning the President's Scholarship from Peking University (PKU). Zhongjie and Nanhao have both been awarded highly competitive postdoctoral fellowships. And Chen was granted tenure. Congratulations to everyone!
AltConf paper online
Congratulations to Jiaxuan on the successful publication of her new paper online! Since March 2019, Jiaxuan has diligently pursued the investigation of whether a deep learning-based approach can effectively generate de novo predictions for multiple state structures from a protein sequence. The outcomes of this research were very promising, leading Jiaxuan to devise a new protocol for modeling the alternative conformations from a given protein structure. Despite encountering a long and intricate review process across various journals, the paper has now been published online in the Journal of Chemical Information and Modeling (JCIM). Special recognition goes to Lei and Zefeng for their invaluable contributions to the paper too.
MM2023 Conference
Chen was invited to give a talk at the Molecular Modelling Conference 2023 (MM2023). This Australasia's premier molecular modelling conference was held at the main campus of the University of Wollongong, Australia, 7-10 Dec 2023, organised by the Association of Molecular Modellers of Australasia (AMMA). Chen talked about our recent series of work on Ca2+ channels.
SARS-CoV-2 paper online
Our collaborative efforts with Prof. Sai Li at Tsinghua University to model and simulate SARS-CoV-2 have been published in Quantitative Biology. Undertaking the challenging task of intricately modeling SARS-CoV-2, we have gained extensive experience in dealing with such complex systems, drawing from our own investigations as well as the research papers of other teams. Dali and Jiaxuan dedicated significant time and effort to modeling the virion structure as accurately as possible, leading to the discovery of interesting dynamics of viral proteins and lipids during coarse-grained Martini simulations. Additionally, we were fortunate to utilize China's E-scale Tianhe-3 supercomputer, conducting all-atom simulations on the massive system comprising 274,193,494 atoms.
Cav paper online
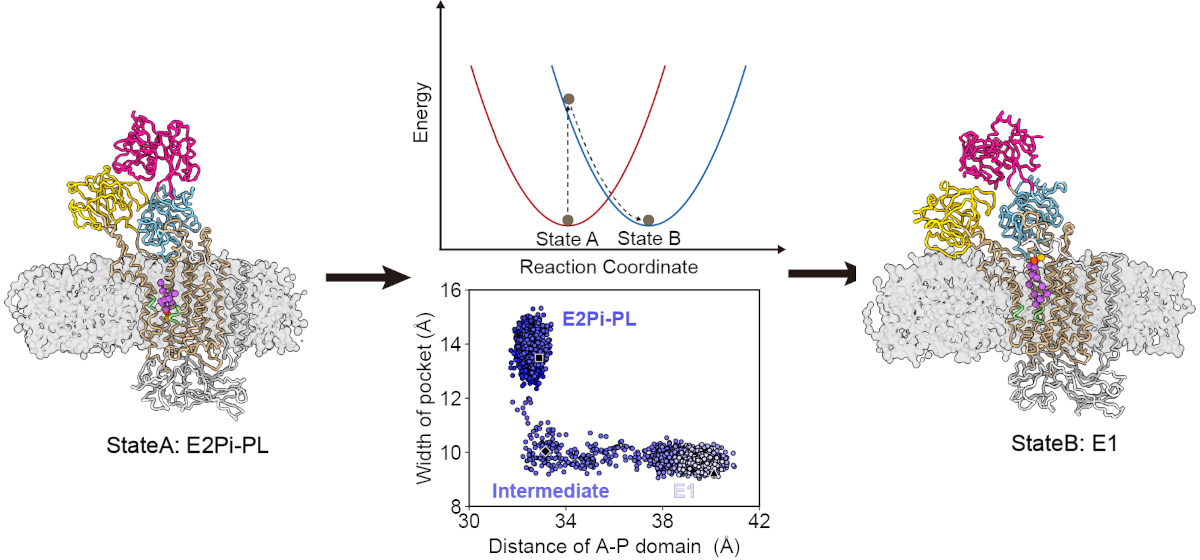
We have been collaborating with Prof. Yan's team to study the structures and functions of Cav1.2, and Lingfeng has done a wonderful job in identifying the functional state of a novel Cav1.2 structure by using all-atom MD simulations. Our collaborative paper has been published in Cell, and we anticipate sharing more interesting stories in the future.