On October 22, 2019, the team of Professor Tang Chao and Assistant Researcher Yang Xiaojing (co-first author: Dr. Qu Yimiao, Dr. Jiang Jun and Dr. Liu Xiang) published 'Cell Cycle Inhibitor Whi5 Records Environmental Information to Coordinate Growth and Division in Yeast’ in Cell Reports. Using budding yeast as a model organism, this paper revealed that cells can record environmental information and regulate cell cycle initiation through Whi5, the key inhibitor of cell cycle initiation.
Cell division mainly includes two processes of genetic materials duplication and cytokinesis. Cell division is the basis for the growth, development and reproduction of eukaryotes and is precisely regulated by highly conserved molecular mechanisms. Cell division usually includes four phases, G1, S, G2 and M phase. During the S and M phases, yeast carries out genetic materials duplication and cytokinesis, respectively. Cells sense their surroundings in the G1 phase, which determines the timing of the initial division. Nutrition, growth factors, pressure signals, oxygen concentration and hormones in the extracellular environment all affect whether and when cell division can start. On the other hand, the timing of cell division initiation, that is, the length of G1 phase, seriously affects whether cell division is normal Complete and whether the cell can survive. But how cells feel complex environmental information, and proper regulation of cell division needs further study.
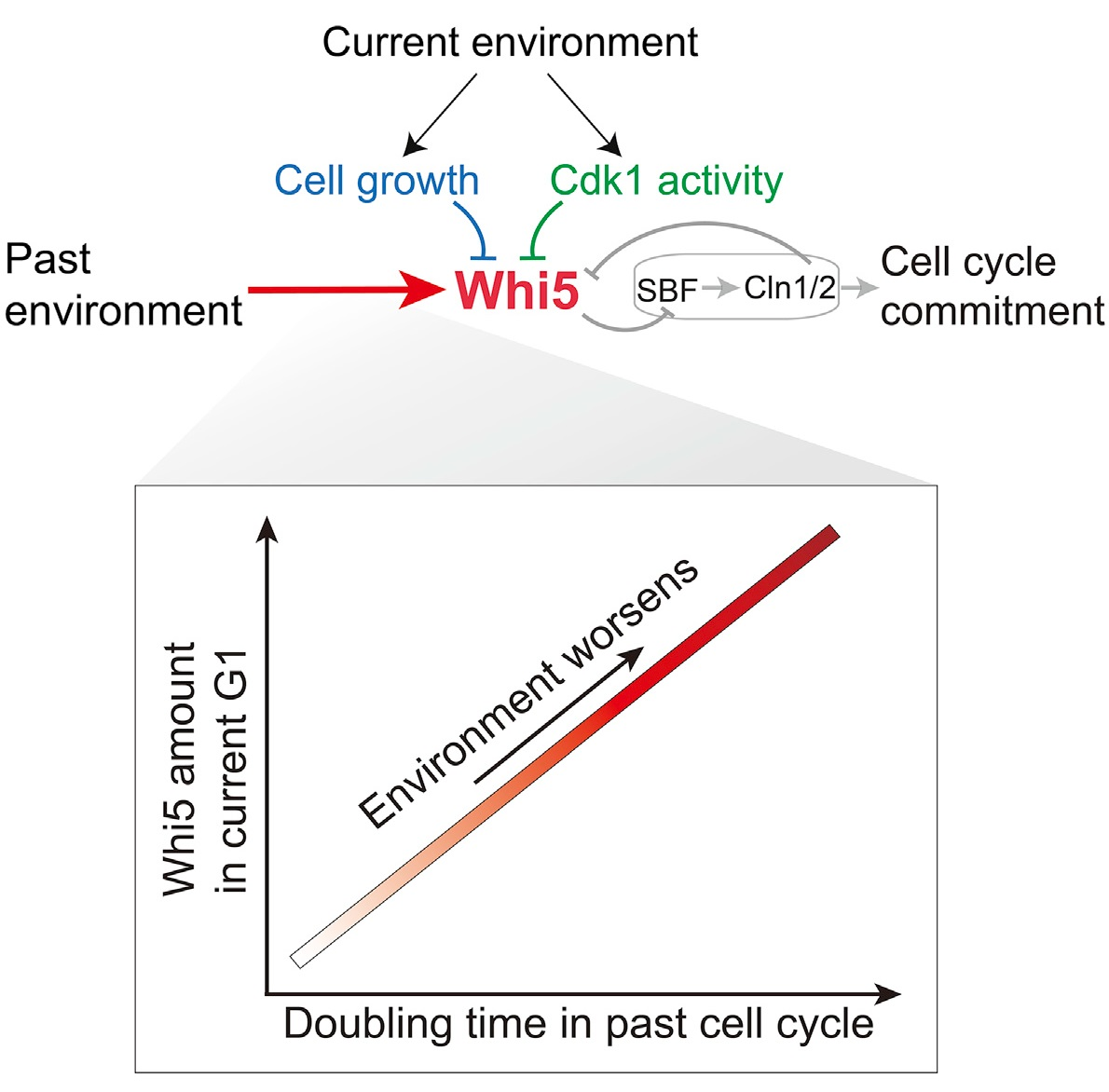
Our article uses budding yeast as a model organism, revealing that cells can record environmental information and regulate cell cycle initiation through Whi5, a key inhibitor of cell cycle initiation. The Whi5 protein binds to and inhibits the key transcription factor SBF that promotes the initiation of the cell cycle in the G1 phase. Inactivation of Whi5 marks the initiation of the cell cycle. We combined fluorescent protein labelling, microfluidic chip technology and microscope timing observation technology to quantify the cell cycle regulation mechanism at the single-cell level. In a variety of different nutrient conditions and stress culture environments, the dynamic of Whi5 levels in single cells with different periods of the cell cycle were tracked, and the cell cycle duration of the corresponding single cells was accurately measured. We found that the nuclear concentration of Whi5 in the early stage of G1 can accurately reflect the duration of the cell cycle, so Whi5 can be used as an indicator that reflects a variety of different environmental conditions.
By measuring the synthesis rate and degradation rate of Whi5 protein from the single-cell level, we further discovered the molecular mechanism of Whi5 protein levels in response to changes in the cell cycle. The average synthesis rate of Whi5 protein in the cell cycle does not change with the environment, and does not degrade during the entire cell cycle, but accumulates over time. Under these mechanisms, Whi5 protein levels are only related to the length of the cell cycle. Therefore, complex environmental information can be quantitatively converted into Whi5 protein levels. Through this mechanism, the Whi5 protein can remember the history of environmental conditions. The budding yeast determines when a new cell division begins based on this quantified environmental information.
Our research found that cells can not only perceive changes in the current environment but also record the past environmental information and quantify them as protein levels of important regulatory factors within the cell. This mechanism is of great significance for new cells to sense the environment and regulate cell division, and coordinate cell division and growth. This discovery reveals a new mechanism of cell cycle regulation and may have a very important reference for studying how cells in a complex environment, such as tumor cells, feel the microenvironment and regulate their proliferation and growth.