As we all know, uranium is the key element used in nuclear energy industry. By many estimates, there are about 4.5 billion tons of uranium dissolved in the world’s oceans—nearly 1,000 times as much of the worldwide reserves of uranium on land—thought to be an inexhaustible supply. Efforts to extract uranyl, the predominant aerobic form of uranium, from seawater date back many decades. So far, though, all solutions have been cost prohibitive. The problem is that the concentration of uranyl is extremely low (3.2 ppb, approximately 14 nM), comparing with other metals with a similar size and charge, which requires the uranyl extraction ligands to be very potent and selective.
Recently, researchers from Professor Luhua Lai’s lab (Center for Quantitative Biology/College of Chemistry and Molecular Engineering, Peking University) and Professor Chuan He’s lab (the University of Chicago) used a computational algorithm to guide the engineering of a protein to selectively bind uranyl with a very high affinity (7.4 fM), which they called it Super Uranyl-binding Protein (SUP). Based on the computational “protein key residues grafting” strategy established by Professor Lai’s lab for protein-protein interaction design, they developed a computational screening algorithm called URANTEIN to search the Protein Data Bank for proteins that could accommodate uranyl. Ten promising candidates were selected, among which four showed binding to uranyl with Kd values of ~100 nM and below. One of the candidates was further optimized with several rounds of amino acid residue mutations, bringing the binding affinity down from 37nM to 7.4 fM (femtomolar) with high selectivity. SUP’s high-resolution crystal structure revealed a binding-site geometry similar to that predicted by the URANTEIN model, proving the rationality of the protein design strategy.Furthermore, the team genetically expressed SUP on the surface of E. coli and found that this bacterial system could extract uranyl selectively from synthetic seawater very efficiently. This is the first known demonstration of a bacterial system used to mine ocean-based uranium, which is cheaper than existing methods. The results were published in Nature Chemistry (Nat. Chem. 2014, 6, 236-241. http://www.nature.com/nchem/journal/v6/n3/full/nchem.1856.html).
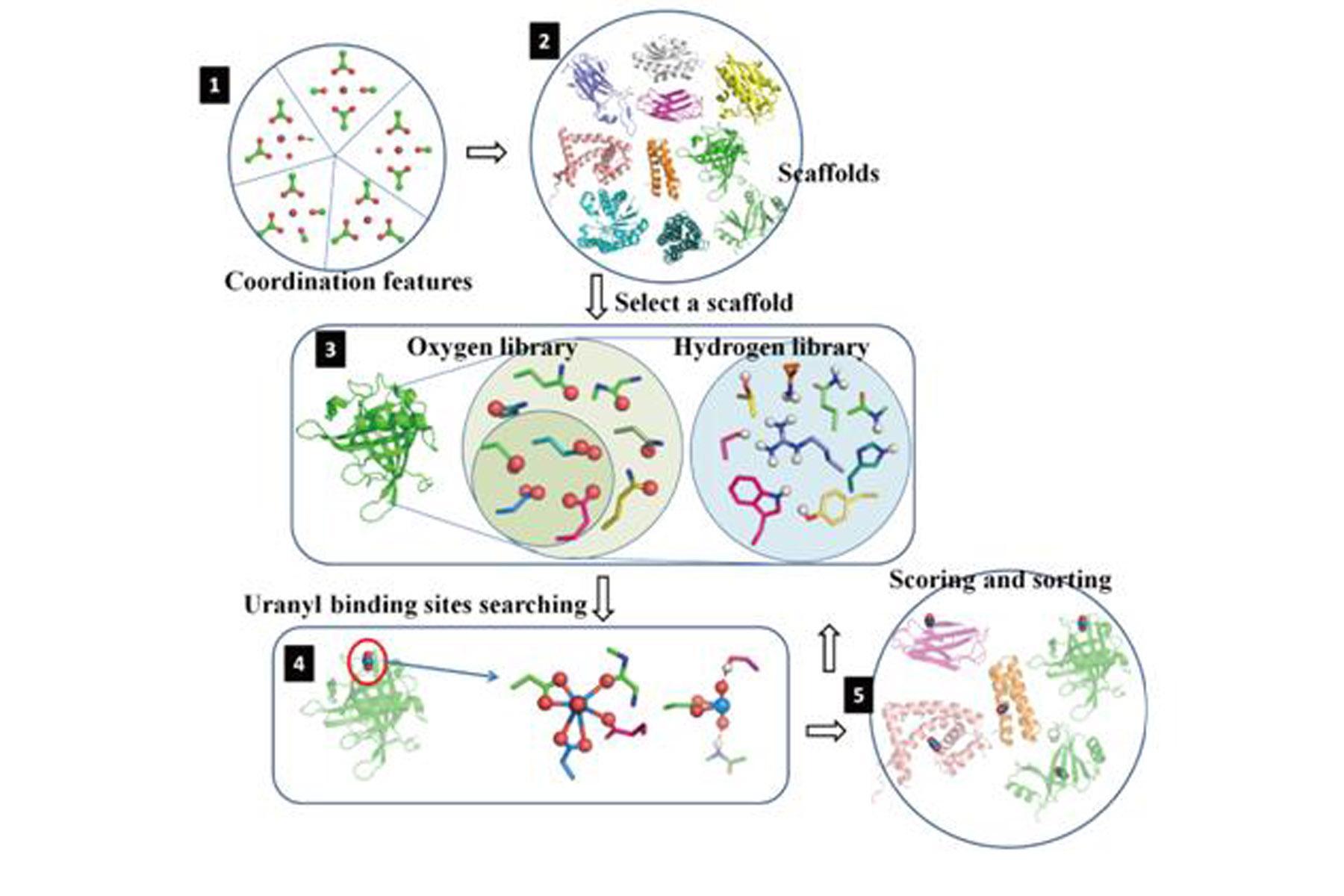
Figure 1. The main steps in computational screening and design uranyl binding proteins
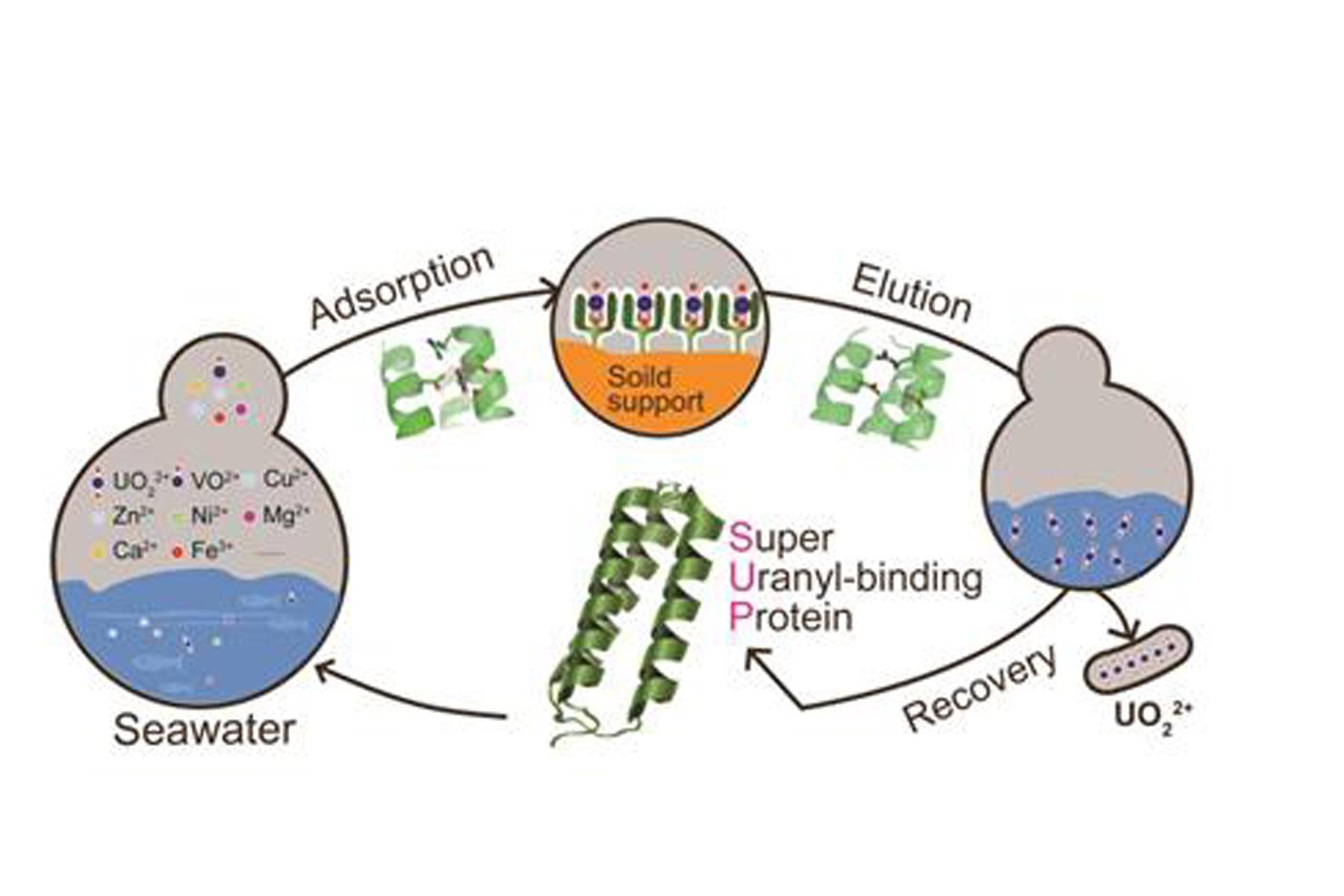
Figure 2. Uranyl sequestration strategy from seawater by Super Uranyl-binding Protein
This work attracted wide attention soon after published online. It was introduced by Chemical & Engineering News (2014, 92, 26) in Science & Technology Concentrates section. It was also highlighted in the same issue of Nat. Chem. (2014, 6, 175), pointing out that: “This work represents not only an important step towards developing efficient strategies to extract uranium from the ocean but also a breakthrough in metalloprotein design.” More recently, it was selected by the management of the Advanced Photon Source (APS) of Argonne National Lab, as one of the outstanding recent results.
Dr. Lu Zhou, Dr. Mike Bosscher from Professor He’s lab and Dr. Changsheng Zhang from Professor Lai’s lab contribute equally to this work. Professor Mark P Jensen from Argonne National Lab also gave strong support on this project. Dr. Lu Zhou, who is currently an associate professor at the School of Pharmacy, Fudan University, worked on this project as a visiting scholar in Professor Chuan He’s lab at Chicago. Dr. Lu Zhou got his B.S. and Ph.D from the College of Chemistry and Molecular Engineering, Peking University.