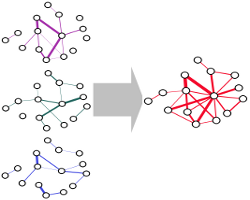
IntNetDB is a database of human protein-protein interactions (PPIs) and their worm, fruitfly and mouse interologs computationally predicted through Bayesian analysis by integrating large-scale physical protein-protein interactions, protein domain assignments, gene co-expression data, genetic interactions, gene context information and biological function annotations from human and other model organisms. Enter >>>
eResponseNet is a package implemented and extended the existing ResponseNet algorithm for prioritizing candidate disease genes through cellular pathways. Using type II diabetes (T2D) as a study case, we demonstrate that eResponseNet outperforms currently available approaches in prioritizing candidate disease genes. More importantly, the package is instrumental in revealing cellular pathways underlying disease-associated genetic variations. Enter >>>
SeqSpider is the first Bayesian network algorithm that enables learning from tag distributions, a unique feature of ChIP-Seq and bisulfite sequencing data, and combined with a profile clustering method for noise removal, enables ab initio identification of interactions from multiple sources of heterogeneous data. Enter >>>
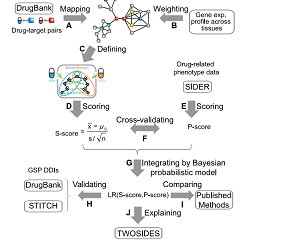
DDI Predictor is a package designed for identifying the drug-drug interaction. Here, we developed a metric S-score that measures the strength of network connection between drug targets to predict PD DDIs. Utilizing known PD DDIs as golden standard positives (GSPs), we observed a significant correlation between S-score and the likelihood a PD DDI occurs. Our prediction was robust.Enter >>>
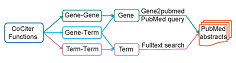
CoCiter is based on literature co-citations to address these disadvantages in conventional gene set and gene function analysis. Co-citation analysis is widely used in ranking articles and predicting protein-protein interactions (PPIs). Our algorithm can further assess the co-citation significance of a gene set with any other user defined gene sets, or to any free terms.Enter >>>
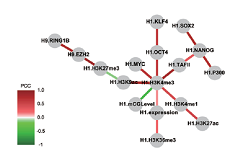
DM_BN is a new bayesian network algorithm. We applied DM_BN to the expression profiles of 544 yeast single or double deletion mutants of transcription factors, chromatin remodeling machinery components, protein kinases and phosphatases in S. cerevisiae. The network inferred by this method identified causal regulatory and non- causal concurrent interactions among these regulators (genetically perturbed genes) that are strongly supported by the experimental evidence, and generated many new testable hypotheses. Compared to networks reconstructed by routine similarity measures or the classical Bayesian network algorithm, the network inferred by DM_BN excels in both accuracy and coverage. Enter >>>
iNPS is an improved algorithm for accurate nucleosome positioning from sequencing data. It achieves significantly better performance than the original NPS, including a significantly higher sensitivity, detection quality, robustness and lower false positive rate in detecting genome-wide nucleosome positions from the MNase-seq data. Further comparison of iNPS with other software packages indicated an overall advantage of iNPS, including higher accuracy with clear nucleosome borders, higher detection coverage with better quality, lower false positive rates, better user convenience, etc. Enter >>>
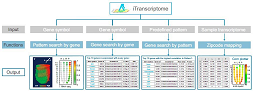
iTranscriptome (for in silico transcriptome) is a web portal to provide open access of the spatial transcriptome data of mouse E7.0 embryo. The iTranscriptome offers data search functionalities in (1) pattern search by gene: querying and displaying the expression pattern of genes of interest in either a corn plot or a digitally reconstructed d-WISH format; (2) gene search by gene: searching for genes that share a similar expression pattern with the queried gene; and (3) search for a set of genes displaying a predefined (20 in the database) or customized (user-defined) expression pattern. Besides these functionalities, the iTranscriptome also enables matching queried cells to their in vivo counterparts in the epiblast by zip code mapping. Enter >>>
1. Guangdun Peng^, Shengbao Suo^, Jun Chen^, Weiyang Chen, Chang Liu, Fang Yu, Ran Wang, Shirui Chen, Na Sun, Guizhong Cui, Lu Song, Patrick P.L. Tam, Jing-Dong J. Han*, Naihe Jing*. Spatial Transcriptome for the Molecular Annotation of Lineage Fates and Cell Identity in Mid-gastrula Mouse Embryo. Developmental Cell, March, 2016.


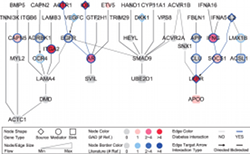
I2CDB is a database collected human datasets involved in the transition process from inflammation to cancer. I2CDB collected different kinds of cancer types such as liver cancer, colon cancer, gastric cancer, esophageal cancer and desmoid cancer. Enter >>>
iCpSc Single-cell RNA sequencing (scRNA-seq) is a powerful method for dissecting intercellular heterogeneity during development. Conventional trajectory analysis provides only a pseu- dotime of development, and often discards cell-cycle events as confounding factors. Here using matched cell population RNA-seq (cpRNA-seq) as a reference, we developed an “iCpSc” package for integrative analysis of cpRNA-seq and scRNA-seq data. By generating a computational model for reference “biological differentiation time” using cell population data and applying it to single-cell data, we unbiasedly associated cell-cycle checkpoints to the internal molecular timer of single cells. Through inferring a network flow from cpRNA-seq to scRNA-seq data, we predicted a role of M phase in controlling the speed of neural differ- entiation of mouse embryonic stem cells, and validated it through gene knockout (KO) experiments. By linking temporally matched cpRNA-seq and scRNA-seq data, our approach provides an effective and unbiased approach for identifying developmental trajectory and timing-related regulatory events.Enter >>>
1. Na Sun^, Xiaoming Yu^, Fang Li^, Denghui Liu, Shengbao Suo, Weiyang Chen, Shirui Chen, Lu Song, Christopher D. Green, Joseph McDermott, Qin Shen, Naihe Jing & Jing-Dong J. Han*. Inference of differentiation time for single cell transcriptomes using cell population reference data. Nature Communications. November, 2017.