Lei and Lingfeng participated in the 2nd National Conference on Biomolecular Structure Prediction and Modeling, held in Hulun Buir, China, from June 27th to 30th, 2024.
Prof. Chen SONG was invited to give a talk at the International Union for Pure and Applied Biophysics (IUPAB2024) congress in Kyoto, Japan , where he presented our recent work titled "Modelling multistate structures of proteins and simulating their conformational transitions". Additionally, members of our group, including Zhongjie, Qingyang, and Haozhe, attended the conference.
The collaborative research on VAChT has been published in Cell Research online! In this paper, we collaborated with Prof. Zhe Zhang's lab and performed molecular dynamics (MD) simulations on the structures of human vesicular acetylcholine transporter (VAChT or SLC18A3) in complex with vesamicol or its native substrate Acetylcholine (ACh). The findings from this work contribute to a better understanding of the structural and functional mechanisms of VAChT and its interactions with two important ligands. Congratulations to Nanhao on the successful publication!
On June 6, 2024, Prof. Frank Noé from the Freie Universität Berlin was invited by Prof. Chen Song to present a CQB seminar titled "Advancing molecular physics with deep learning". In his presentation, Prof. Frank Noé provided an overview of highly accurate computational methods for determining quantum states using deep fermionic neural networks and Quantum Monte Carlo. He also discussed the approaches to address the many-body sampling problem with generative deep learning.
We are excited to welcome Qiushi and Jun to our group after they completed their three rotations! Qiushi will focus on the rational design of antimicrobial peptides, while Jun will work on representations of protein dynamics using protein pre-trained models in the future. Their diverse expertise will undoubtedly enhance our research efforts, and we look forward to their contributions.
Invited by Prof. Chen Song, Prof. Wonpil lm from Lehigh University visited us and presented his groundbreaking work on the biomolecular simulation program CHARMM-GUI. CHARMM-GUI is a widely-used software suite that facilitates the setup, simulation, and analysis of a diverse range of biomolecular systems, including proteins, lipids, and nucleic acids. In his presentation, Prof. Wonpil lm highlighted the exceptional capabilities and versatile applications of this powerful computational biology tool, which has become an invaluable resource for the global research community.
The collaborative project focused on SLC19A3 has been published in Cell Research online! The study was conducted in partnership with Prof. Zhe Zhang's lab, and we employed molecular dynamics (MD) simulations and binding free energy calculations to help determine the complex structures of human thiamine transporter 2 (ThTr2 or SLC19A3). Congratulations to Nanhao on the successful publication of this research!
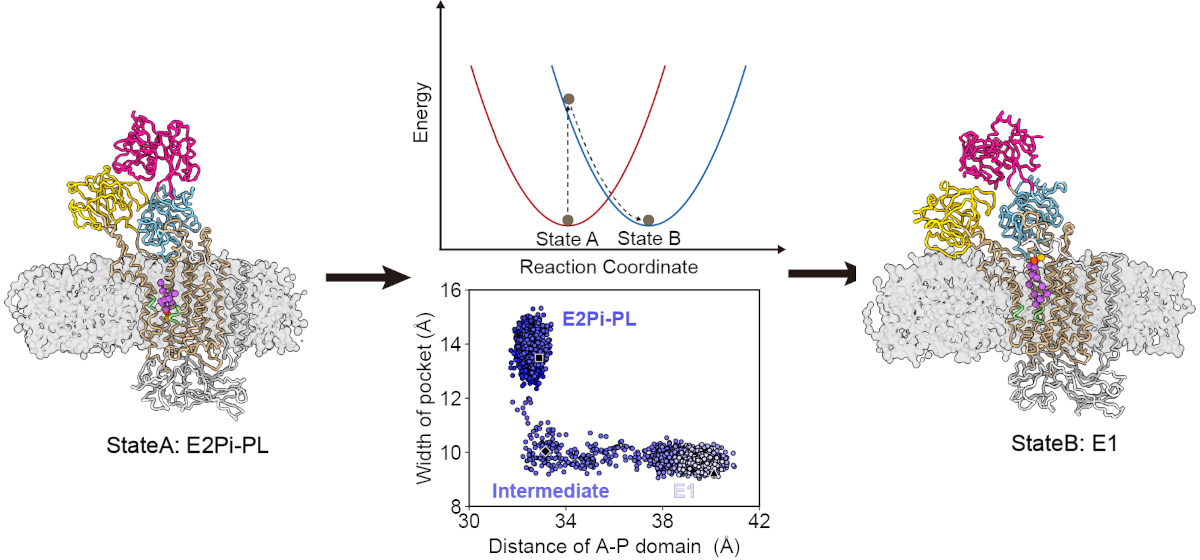
YANG Song's first paper detailing the development of the Switching Gō-Martini method has been published online in JCTC (Link). This method promises to be a valuable tool for efficiently simulating the likely conformational transitions between two structures of a protein. When used in conjunction with Jiaxuan's Alternative Conformation Modeling approach (Link), in principle, one can begin with a protein sequence, predict two probable conformations, and then simulate the potential transition pathways between them.